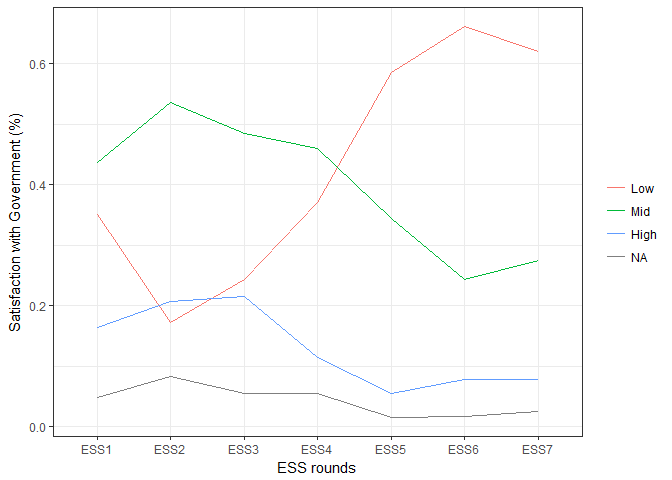
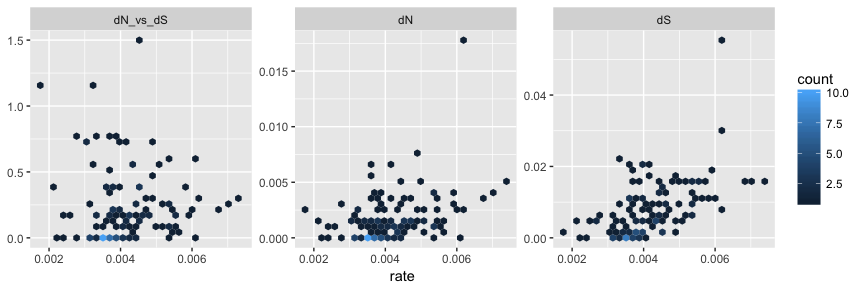
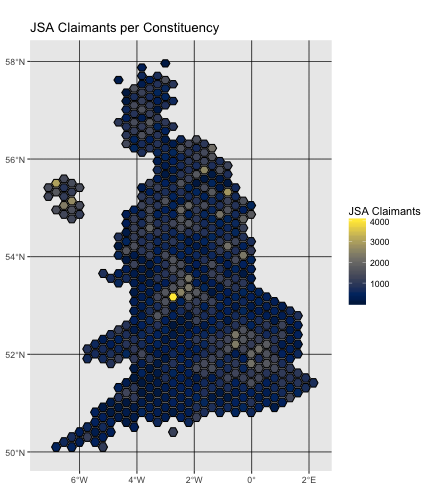
Evolutionary biologists are increasingly using R for building,editing and visualizing phylogenetic trees.The reproducible code-based workflow and comprehensive array of toolsavailable in packages such as ape,phangorn andphytools make R an ideal platform forphylogenetic analysis.Yet the many different tree formats are not well integrated,as pointed out in a recentpost.