You can find members of the rOpenSci team at various meetings and workshops around the world. Come say ‘hi’, learn about how our packages can enable your research, or about our onboarding process for contributing new packages, discuss software sustainability or tell us how we can help you do open and reproducible research.Where’s rOpenSci?
Since June, we have been highlighting the many projects that emerged from this year’s rOpenSci Unconf. These projects start many weeks before unconf participants gather in-person. Each year, we ask participants to propose and discuss project ideas ahead of time in a GitHub repo. This serves to get creative juices flowing as well as help people get to know each other a bit through discussion.
elastic is an R client for Elasticsearch elastic has been around since 2013, with the first commit in November, 2013.What is Elasticsearch? If you aren’t familiar with Elasticsearch, it is a distributed, RESTful search and analytics engine.It’s similar to Solr. It falls in the NoSQL bin of databases, holding data in JSON documents, insteadof rows and columns.
How do you get the maximum value out of a dataset? Data is most valuable when it can easily be shared, understood, and used by others. This requires some form of metadata that describes the data. While metadata can take many forms, the most useful metadata is that which follows a standardized specification. The Ecological Metadata Language (EML) is an example of such a specification originally developed for ecological datasets.
What is Taxonomy? Taxonomy in its most general sense is the practice and science of classification. It can refer to many things. You may have heard or used the word taxonomy used to indicate any sort of classification of things, whether it be companies or widgets. Here, we’re talking about biological taxonomy, the science of defining and naming groups of biological organisms.

Most of us who work in R just want to Get Stuff Done™. We want a minimum amount of friction between ourselves and the data we need to wrangle, analyze, and visualize. We’re focused on solving a problem or gaining insights into a new area of research. We rely on a rich, community-driven ecosystem of packages to help get our work done and likely make an unconscious assumption that there is a safety net out there, protecting us from harm.

In my training as a AAAS Community Engagement Fellow, I hear repeatedly about the value of extending a personal welcome to your community members. This seems intuitive, but recently I put this to the test. Let me tell you about my experience creating and maintaining a #welcome channel in a community Slack group.
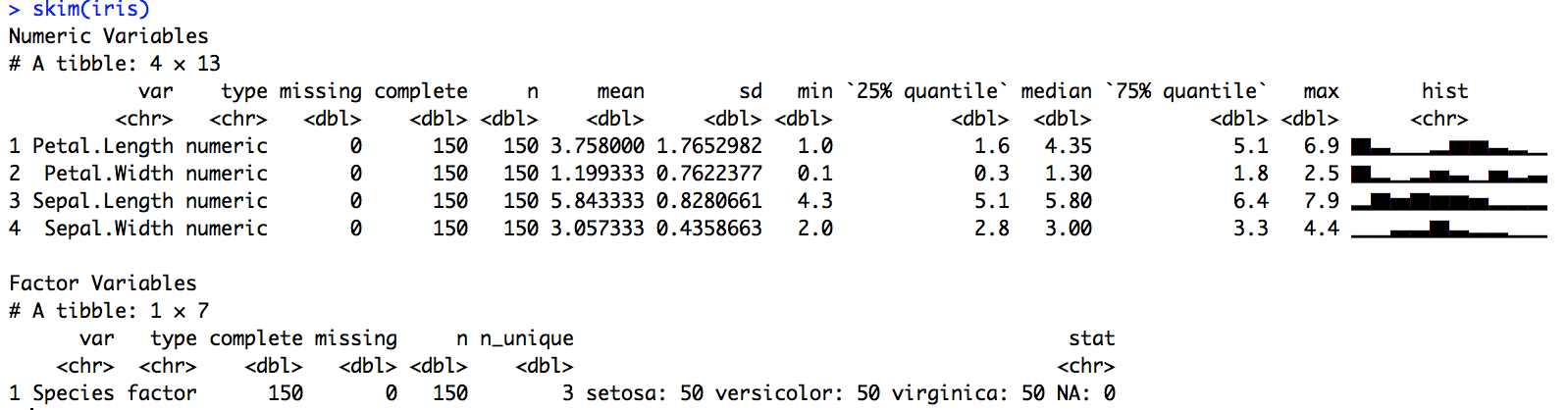
Like every R user who uses summary statistics (so, everyone), our team has to rely on some combination of summary functions beyond summary() and str(). But we found them all lacking in some way because they can be generic, they don’t always provide easy-to-operate-on data structures, and they are not pipeable. What we wanted was a frictionless approach for quickly skimming useful and tidy summary statistics as part of a pipeline.
rOpenSci’s mission is to promote a culture of open, transparent, and reproducible research across various research domains. Everything we do, from developing high-quality open-source software for data science and, software review, to building community through events like our community calls and annual unconference are all geared toward lowering barriers to reproducible, open science.
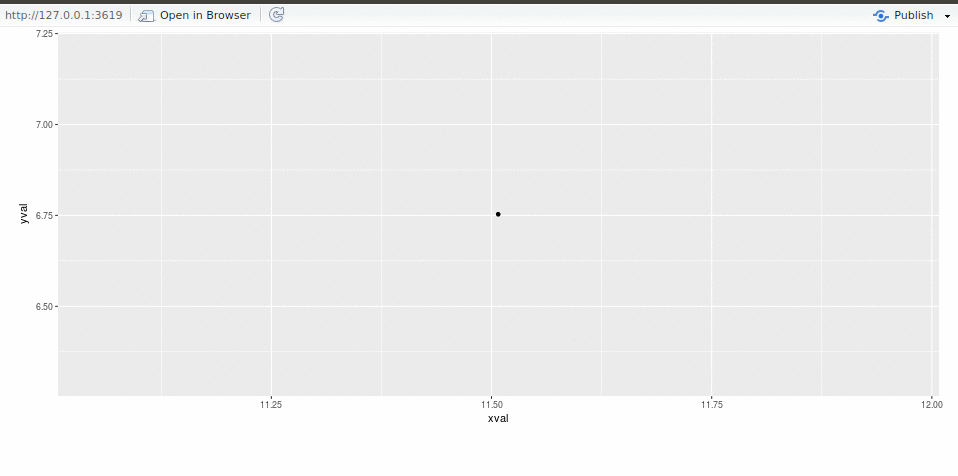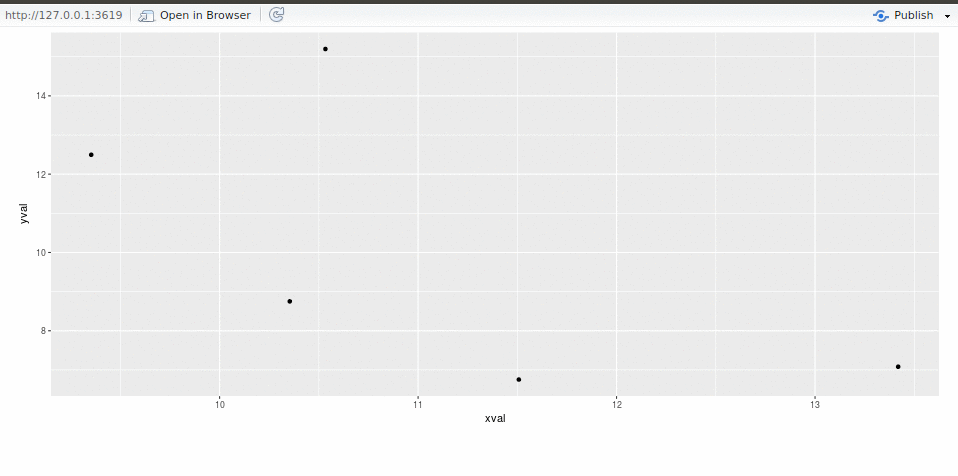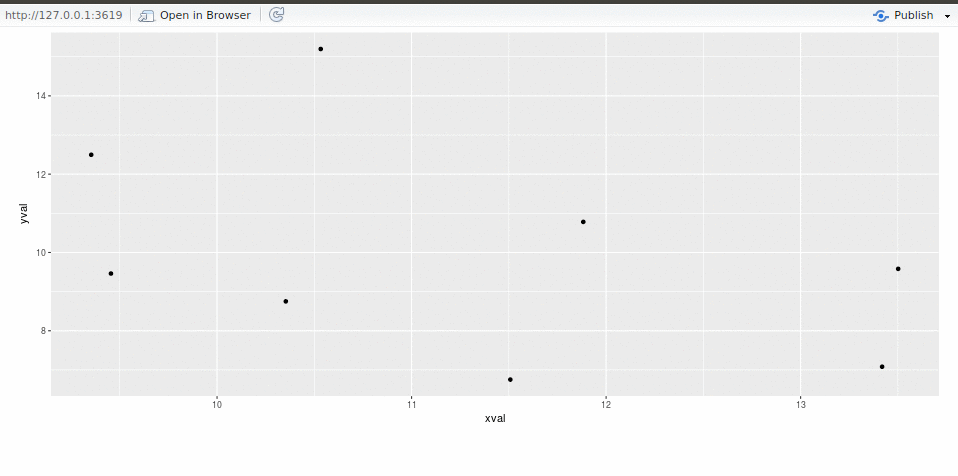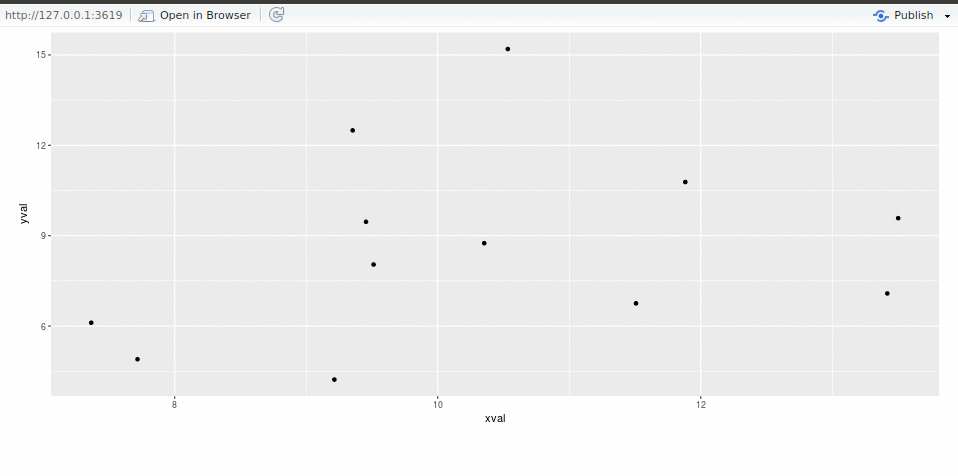
We, Alicia Schep and MilesMcBain, drove the webrockets projectat #runconf17.To make progress we solicited code, advice, and entertaining anecdotesfrom a host of other attendees, whom we humbly thank for helping to makeour project possible. This post is divided into two sections: First up we’ll relate ourexperiences, prompted by some questions we wrote forone another.