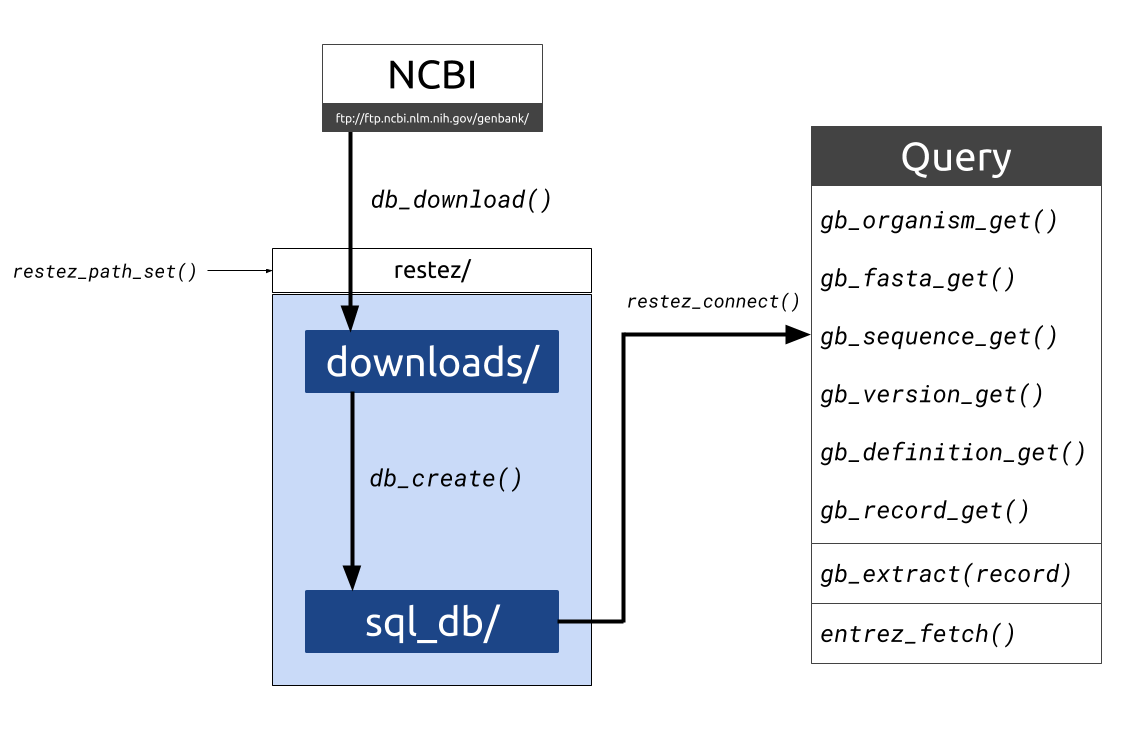
🎤 Dan Sholler, rOpenSci Postdoctoral Fellow 🕘 Tuesday, December 18, 2018, 10-11AM PST; 7-8PM CET (find your timezone) ☎️ Details for joining the Community Call. Everyone is welcome. No RSVP needed. Researchers use open source software for the capabilities it provides, such as streamlined data access and analysis and interoperability with other pieces of the scientific computing ecosystem.