I have written previously about Journal Impact Factors (here and here). The response to these articles has been great and earlier this year I was asked to write something about JIFs and citation distributions for one of my favourite journals. I agreed and set to work. Things started off so well. A title came straight to mind.
Yesterday I tried a gedankenexperiment via Twitter, and asked: If you could visualise a protein relative to an intracellular structure/organelle at ~5 nm resolution, which one would you pick and why? https://twitter.com/clathrin/status/707949738323218432 I got some interesting replies: Myosin Va and cargo on actin filaments in melanocytes – Cleidson Alves @cleidson_alves COPII components relative to ER and Golgi for export of

There have been calls for journals to publish the distribution of citations to the papers they publish (1 2 3). The idea is to turn the focus away from just one number – the Journal Impact Factor (JIF) – and to look at all the data. Some journals have responded by publishing the data that underlie the JIF (EMBO J, Peer J, Royal Soc, Nature Chem). It would be great if more journals did this.

A few days ago, Retraction Watch published the top ten most-cited retracted papers. I saw this post with a bar chart to visualise these citations. It didn’t quite capture what the effect (if any) a retraction has on citations. I thought I’d quickly plot this out for the number one article on the list. The plot is pretty depressing. The retraction has no effect on citations.
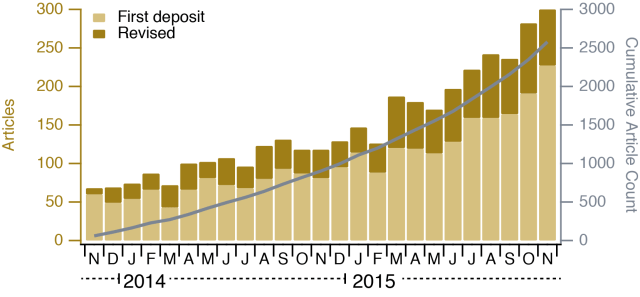
bioRxiv, the preprint server for biology, recently turned 2 years old. This seems a good point to take a look at how bioRxiv has developed over this time and to discuss any concerns sceptical people may have about using the service.
Our recent paper on “the mesh” in kinetochore fibres (K-fibres) of the mitotic spindle was our first adventure in 3D electron microscopy. This post is about some of the new data analysis challenges that were thrown up by this study. I promised a more technical post about this paper and here it is, better late than never.

In the lab we have been doing quite a bit of analysis of cell migration in 2D. Typically RPE1 cells migrating on fibronectin-coated glass. There are quite a few tools out there to track cell movements and to analyse their migration. Naturally, none of these did quite what we wanted and none fitted nicely into our analysis workflow. This meant writing something from scratch in IgorPro. You can access the code from my GitHub pages.
I’m putting this up here in case it is useful for somebody. We capture Z-stacks on a Perkin Elmer Spinning Disk microscope system. I wanted to turn each stack into a single image so that we could quickly compare them. This simple macro does the job. We import the images straight from the *.mvd2 library using the wonderful BioFormats import tool. We open all files as composite hyperstacks.

We have a new paper out! You can access it here. Title of the paper: The mesh is a network of microtubule connectors that stabilizes individual kinetochore fibers of the mitotic spindle What’s it about? When a cell divides, the two new cells need to get the right number of chromosomes. If this process goes wrong, it is a disaster which may lead to disease e.g. cancer.

I was searching for the excitation and emission spectra for mNeonGreen. I was able to find an image, but no values for the spectra.
