The following poem by David Maddison was published in Systematic Biology (doi:10.1093/sysbio/sys057) under a CC-BY-NC license.I think that I shall never seeA thing so awesome as the TreeThat links us all in paths of genesDown into depths of time unseen;Whose many branches spreading wideHouse wondrous creatures of the tide,Ocean deep and mountain tall,Darkened cave and waterfall.Among the branches we may findCreatures there of every
iPhylo
Quick test of an idea I'm playing with.
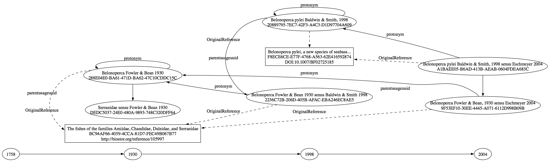
I'm trying to get my head around the data model used by ZooBank to store taxonomic names. To do this, I've built a graph for the species Belonoperca pylei described by Baldwin &
Quick note to self about possible way to using fuzzy matching when searching for taxonomic names. Now that I'm using Cloudant to host CouchDB databases (e.g., see BioStor in the the cloud) I'd like to have a way to support fuzzy matching so that if I type in a name and misspelt it, there's a reasonable chance I will still find that name. This is the "did you mean?" feature beloved by Google users.

Quick note on an experimental version of BioStor that is (mostly) hosted in the cloud. BioStor currently runs on a Mac Mini and uses MySQL as the database. For a number of reasons (it's running on a Mac Mini and my knowledge of optimising MySQL is limited) BioStor is struggling a bit.

Benoît Fontaine et al. recently published a study concluding that average lag time between a species being discovered and subsequently described is 21 years.The paper concludes:This is a conclusion that merits more investigation, especially as the title of the paper suggests there is an appalling lack of efficiency (or resources) in the way we decsribe biodiversity.

CrossRef have released CrossRef Metadata Search a nice tool that can take a free-form citation and return possible matches from CrossRef's database. If you get a match CrossRef can take the DOI and format for you it in a variety of styles using DOI content negotiation.If, like me, you spend a lot of time trying to find DOIs (and other identifiers) for articles by first parsing citations into their component parts, then this is good news.
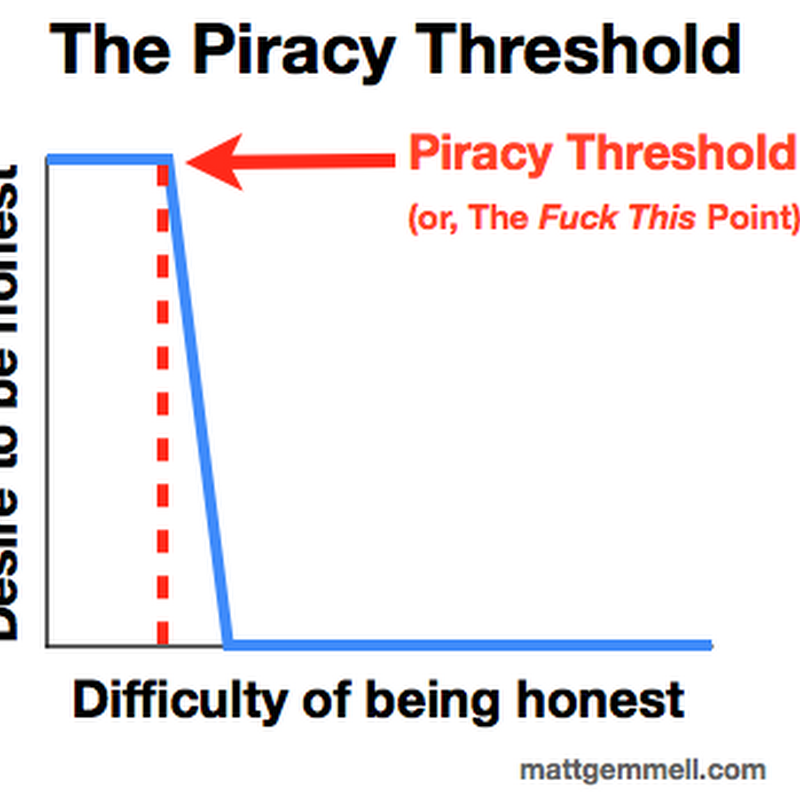
It is well known that phylogeny databases such as TreeBASE capture a small fraction of the published phylogenies. This raises the question of how to increase the number of trees that get archived.

James Rosindell's OneZoom tree viewer is out and the paper describing the viewer has been published in PLoS One (disclosure, I was a reviewer):Below is a video where James describes OneZoom.OneZoom is fun, and is deservedly attracting a a lot of attention. But as visually striking as it is, I confess I have reservations about fractal-based viewers. For a start they make it hard to get a sense of the relative size of taxonomic groups.

Robert M. Griffin (@GriffinEvo) has launched ProjectEvoMap. Rob explains:Below is a screen shot of part of the map.