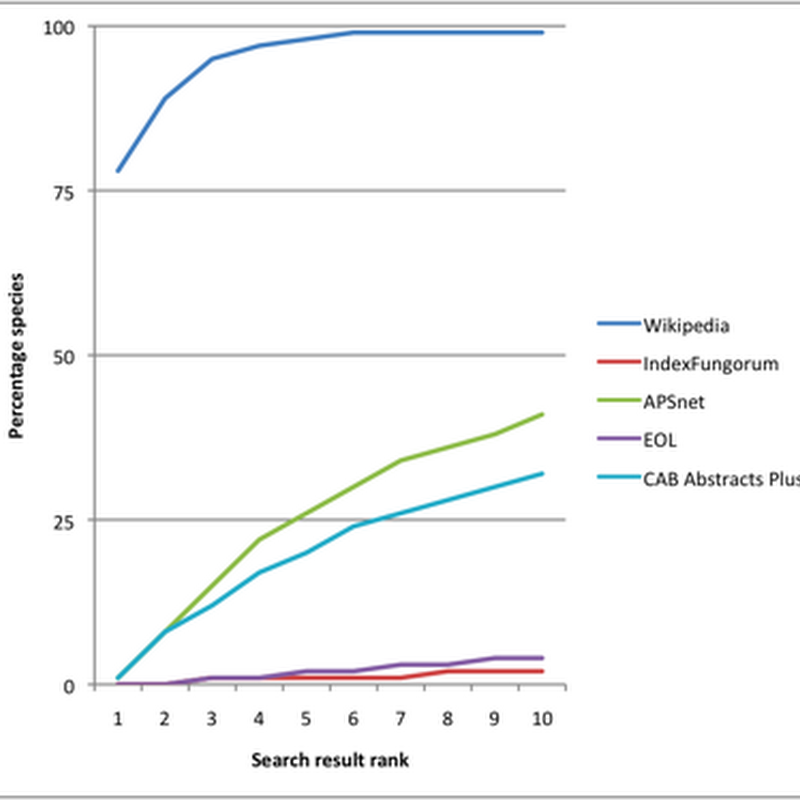
Quick post (really should be doing something else). Reading Jeff Atwood's post Mixing Oil and Water: Authorship in a Wiki World lead me to IBM's wonderful history flow tool to visualise the edit history of a Wikipedia page. There's a nice paper describing history flow (doi:10.1145/985692.985765, free PDF here). Inspired by this I decided to try and implement history flow in PHP and SVG.