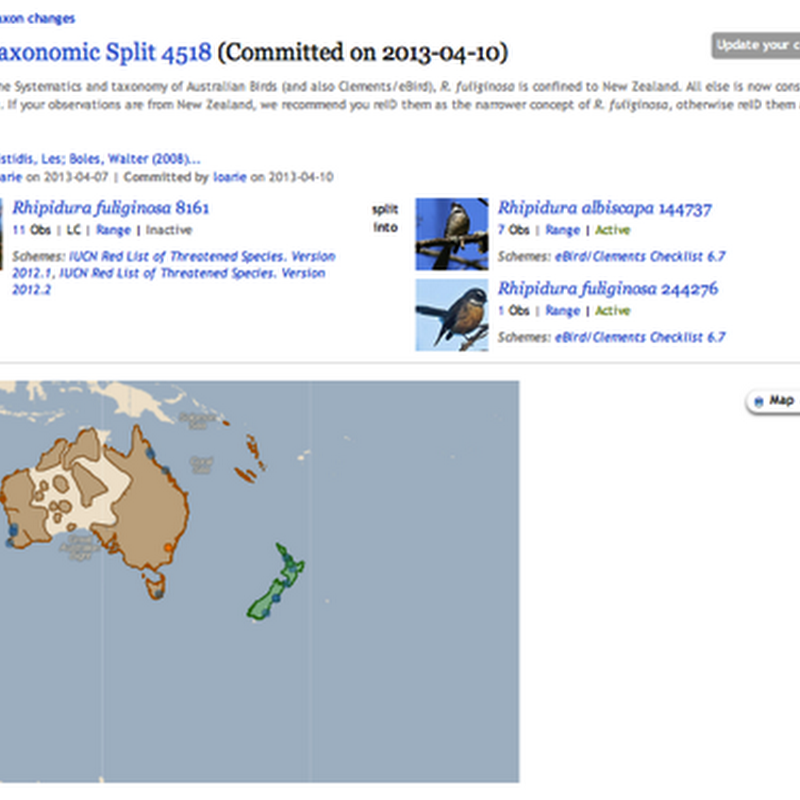
Donald Hobern drew my attention to nice the way iNaturalist displays taxonomic splits: In this example, observations identified as Rhipidura fuliginosa are being split into Rhipidura fuliginosa and Rhipidura albiscapa . This immediately reminds me of the idea which keeps circulating around, namely using version control tools to manage taxonomic classification.




